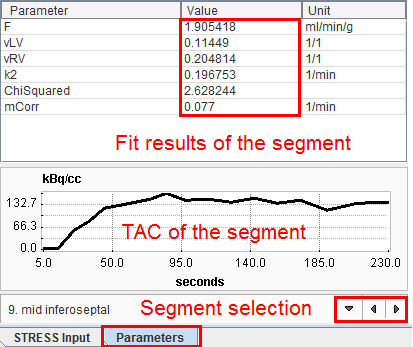
Parameters per Segment
The Parameters tab on the Kinetic Modeling page shows the modeling results obtained with the individual segmental TACs. After a segment has been selected, the result parameters are shown in the upper table, and the corresponding TAC in the lower curve area.
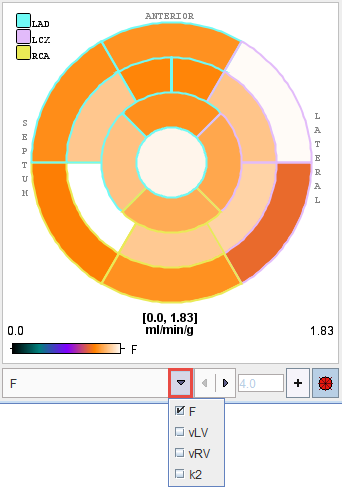
Polar Plots
The parameters resulting in the different segments can conveniently be visualized as polar plots. The example below shows the polar plot of the parameter flow (F) , without interpolation and with the display controls enabled. By default, segments definition is overlayed. These can be easily switched off selecting the toggle button ![]() available on the lower right corner of the polar plot. The + button serves for defining a standard maximal value, which can be entered in the number field left to it.
available on the lower right corner of the polar plot. The + button serves for defining a standard maximal value, which can be entered in the number field left to it.
A list selection is available at the bottom to switch between the parameters. When the cursor is moved about the polar plot the instantaneous value is shown in the upper right. Note that the segment structure is clearly visible if no interpolation has been configured for the polar plot. The polar plot can be documented by the Polar Plot report.
The polar plot display control can be easily switch on/off right clicking on the polar plot area and selecting the Hide/Show Controls option.
Activating the last option on the list, Properties, the display properties like Description's font size can be changed.
Resampled Data and Parameter Images
The image display on the right side of Kinetic Modeling page initially shows the MYOCARDIUM data. Note, however, that you can switch between more studies by the list selection as illustrated below. Green bullets represent data already available, while red bullets mark data which still needs to be calculated (in the example modeling has not yet been performed).
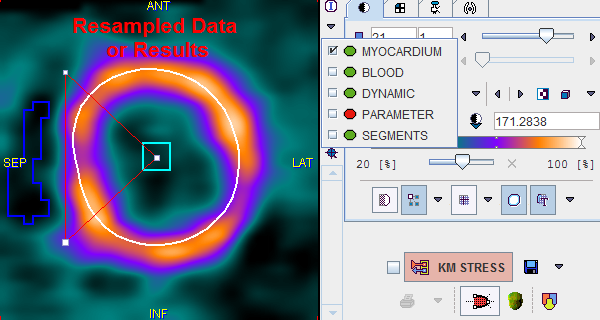
All this data has been resampled based on the Myocard VOI, so that the 20 slices cover the area from the apex to the base, plus some additional bounding box.
DYNAMIC |
Resampled dynamic image data. Use the volume slider to step through the different acquisition times. |
BLOOD |
The data calculated in the pre-processing step as a representation of blood. |
MYOCARDIUM |
The data calculated in the pre-processing step as a representation of myocardium (already illustrated above). |
PARAMETER |
The calculated active parameter (eg. flow F) turned into a volumetric image (one value per segment). The interpolation has been disabled in the illustration above to clearly demonstrate the segmental parameter values. Note that the result images can be saved, for instance for fusion with a matched anatomical data set. |
SEGMENTS |
Illustration of the segments. Note: The representation is based on the myocardium contour. The actually sampled voxels may differ to some extent depending on the sampling scheme. |
Note: If you click into the polar plot, the image to the right is adjusted so that the corresponding slice is shown. With an orthogonal image layout, the cross is positioned at the sampling location corresponding to the polar plot point.
Visualization of the Samples found by Polar Sampling (3D Option required)
The pixels found during the sampling process can be visualized together with the short axis images using the buttons

The left button enables the sampling point visualization. The right button calls the 3D tool which renders the shape of the myocardium model together with the sampling points. The example below shows the sampling points as red spheres, the myocardium ENDO and EPI models as a surfaces with the texture of the MYOCARDIUM image, and a short axis slice of the anatomical images with transparency.
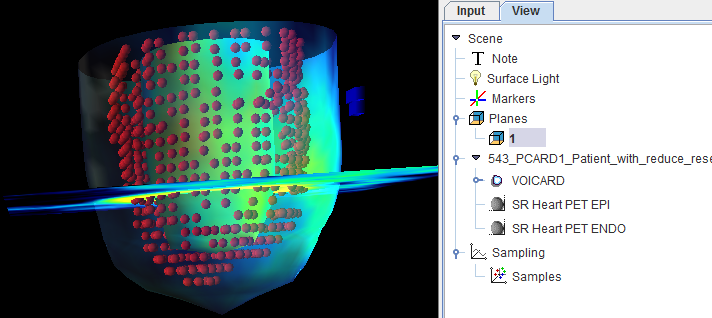
If a sampling scheme with averaging has been selected, the neighbor points are rendered as separate sphere objects in green as illustrated below.
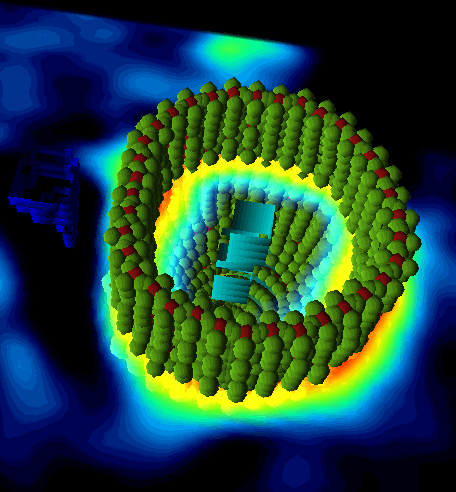
To learn more about the many possibilities exploring the data in P3D please refer to the PMOD 3D Rendering User Guide.